Lattice Nucleoids: Coarse-grain Modeling of Genomes and Associated Proteins
Our lattice nucleoid modeling methods are designed to create models of entire genomes with a manageable computational effort, allowing exploration of multiple hypotheses. The models are created in two steps: first, a lattice-based method is used to trace the overall path of DNA within the given space of a bacterium or virion, and second, the lattice model is relaxed to conform to properties of DNA and interactions with proteins, messenger RNA, and ribosomes.
These methods are currently under active development, so separate versions of the method have been implemented for specific applications. Two versions are currently available:
- Lattice models of bacterial nucleoids
- Lattice models of maturation of the HIV-1 nucleoid
You can send your impressions to me at goodsell@scripps.edu.
This work has been kindly supported by the US National Institutes of Health R01-GM120604 and P50-GM103368 to the HIVE Center.
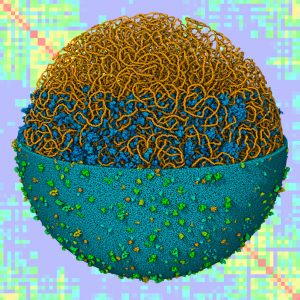
Model of a mycoplasma with lattice-modeled DNA, overlaid on simulated Hi-C data.