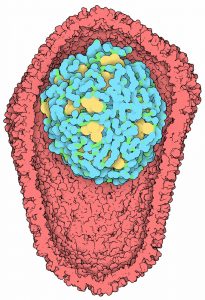
Lattice Models of HIV-1 Nucleoid Condensation
Introduction
Four programs are included in this release, which together model the mature HIV-1 nucleoid, using this five-step pipeline:
1) quasisymmetry.f — creates a lattice of points on a sphere that correspond to the locations of gag proteins in the immature HIV-1 virion
2) rnapath.f — performs a self-avoiding random walk on this lattice, generating the path of two HIV-1 gRNA strands.
3) condense.f — in a short optimization run, is used to relax the RNA path based on a 3 nt/bead coarse-grain model.
4) makeconstraints.f — assigns the location of integrase and nucleocapsid proteins, based on a list of experimental
localizations and choosing integrase sites that are in proximity from step (3).
5) condense.f — condenses the nucleoid incorporating integrase and nucleocapsid localizations and user-defined secondary structure constraints.
The result is a PDB-format file that may be read by most molecular graphics programs. The PDB format is modified in the following ways:
–coordinates are in nanometers
–RNA strands have atom name “P” and residue name “RNA”
–integrase subunits have atom name “CA” and residue name “IN”
–nucleocapsid subunits have atom name “N”, and residue names
“NCO” for experimentally-observed positions and
“NCR” for randomly-placed positions
Suggested radii for visualization: RNA=0.5, NC=1.5, IN=2.5
To create models:
1) Compile the programs:
gfortran quasisymmetry.f -o quasisymmetry
gfortran rnapath.f -o rnapath
gfortran makeconstraints.f -o makeconstraints
gfortran condense.f -o condense
2) The programs and pipeline require several input data files:
icos.mat — matrices for icosahedral symmetry
Tlattice.pdb — coordinates for quasisymmetrical tiling
inpeaks.dat — experimental integrase binding sites
ncpeaks.dat — experimental nucleocapsid binding sites
5UTR.const — constraint file for secondary structure in the 5’UTR
3a) The programs may be run individually, reading input commands from Unit 5, or from a command file, such as:
./condense < condense.inp > condense.log (log file is optional)
3b) The script file HIVnucleoid.script will run a full pipeline to generate a nucleoid model with integrase tetramers.
./HIVnucleoid.script > HIVnucleoid.log (log file is optional)
On typical desktop hardware, this will require about 1/2 hour.
The script file HIVnucleoid_3types.script will run a pipeline to generate three models: with tetramers, dimers, or no integrase.
Please note that these programs were created to explore a specific application, and are not designed for generalized use. They have very little error checking, so large changes in input parameters will be expected to lead to spurious results or program crashes.