This page was last updated on October 17th, 2019 at 11:24 pm
MSMS home page
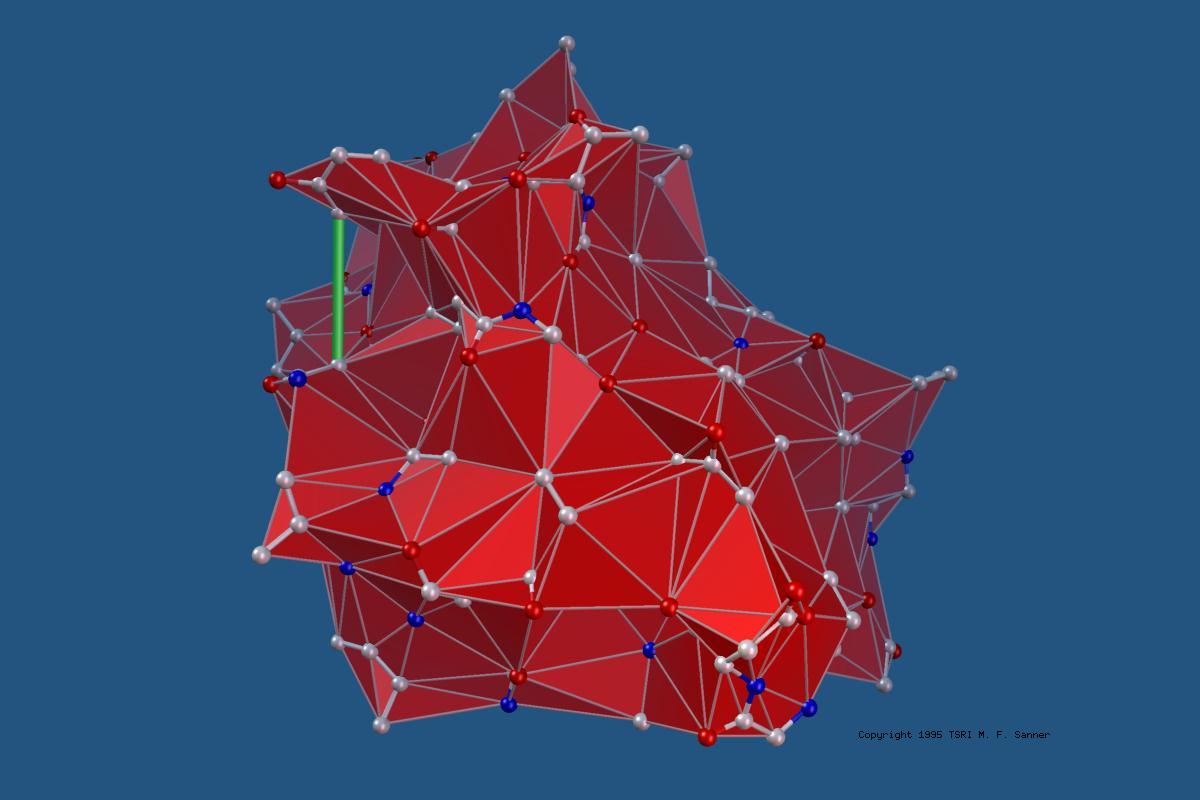
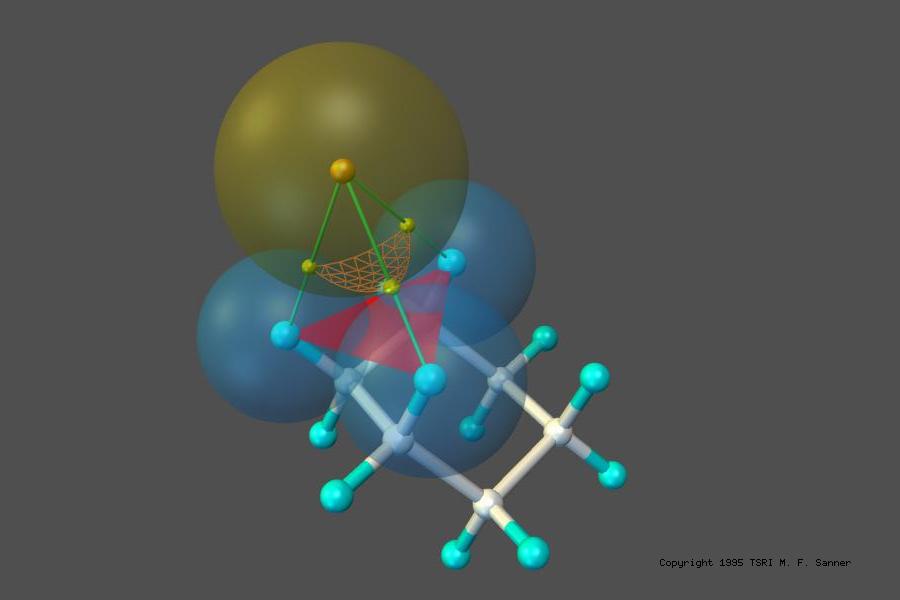
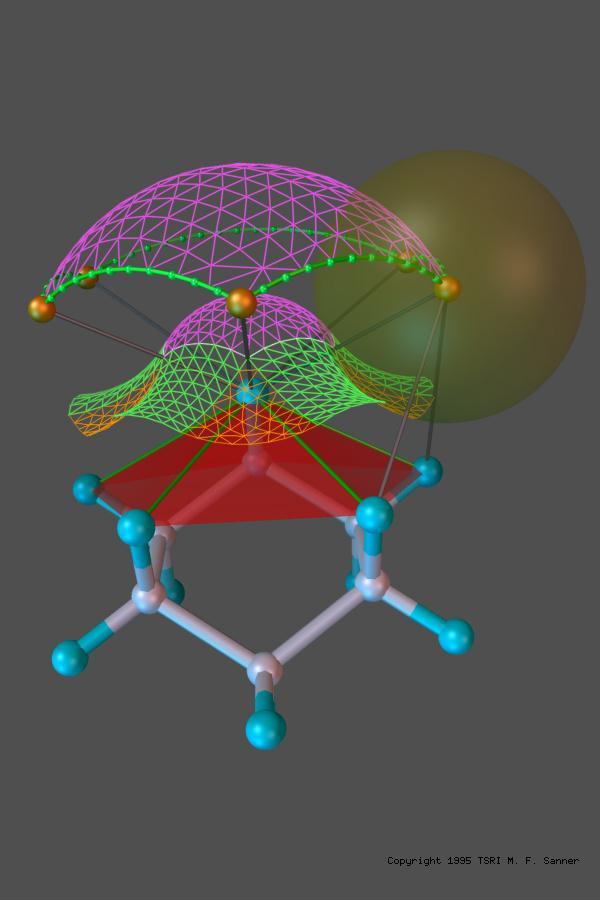
MSMS is a fast algorithm for computing molecular surfaces. It was developed by Dr. Michel F. Sanner as part of his PhD thesis. It has been widely used for computing and displaying Solvent Excluded Surfaces for proteins also knows as the Lee and Richards (who first defined the concept of rolling a probe representing a solvent molecule over the set of sphere representing protein atoms, or the Connolly surface as Mike Connolly wrote the first implementation of a program to compute this surface analytically. MSMS is used by various molecular visualization programs including VMD, Chimera, and our own PMV. It is also available as a standalone binary on the download page as well as a library with Python binding available it the MGLTools and ADFRsuite software distributions.
If you use MSMS for your work please so kind and cite the following paper:
Sanner, M. F., Olson A.J. & Spehner, J.-C. (1996). Reduced Surface: An Efficient Way to Compute Molecular Surfaces. Biopolymers 38:305-320.