This page was last updated on April 25th, 2019 at 10:23 pm
AutoGridFR home page
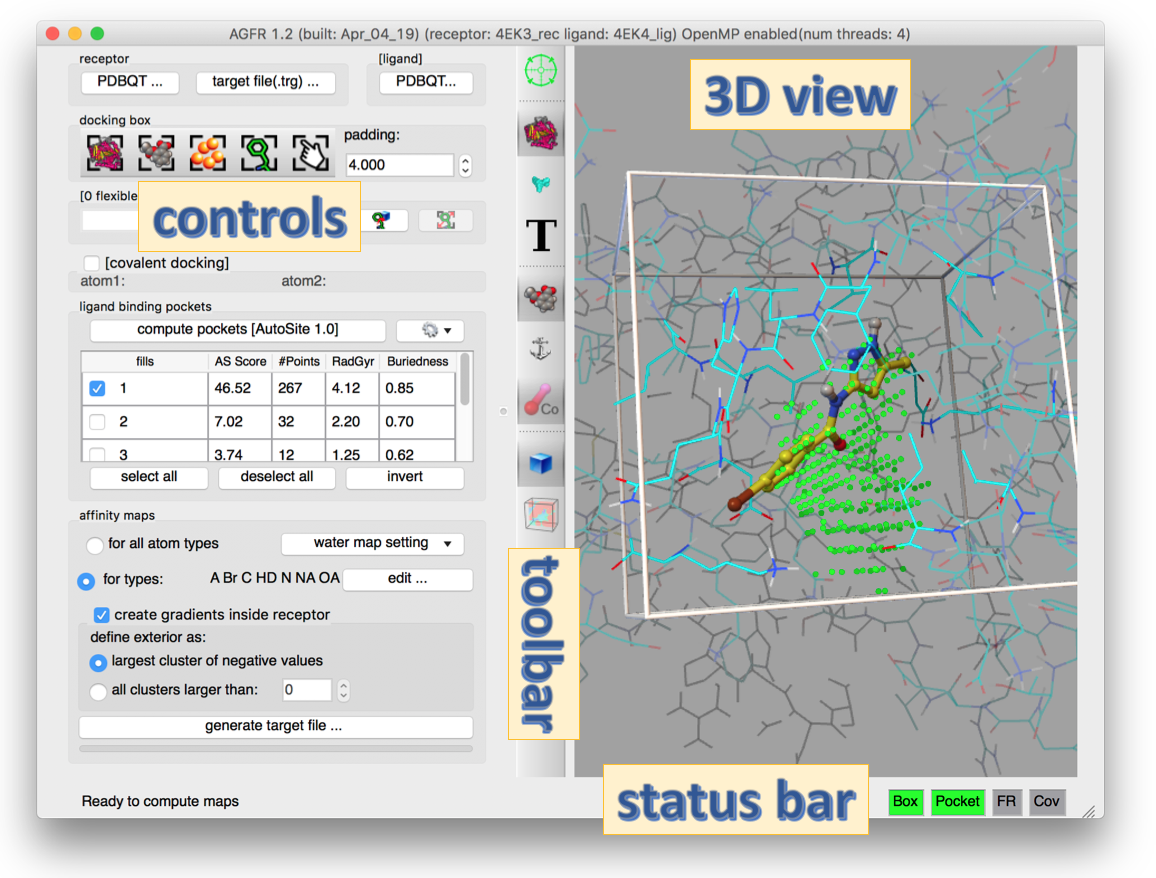
AutoGridFR (or AGFR in short) is a software program that facilitates the calculation of affinity maps used by AutoDockFR, AutoDock CrankPep, and AutoDock4 to represent the binding sites in biological molecules we call receptors in the context of automated docking. AGFR is included in the ADFR software suite available for Linux, Mac OS and Windows.
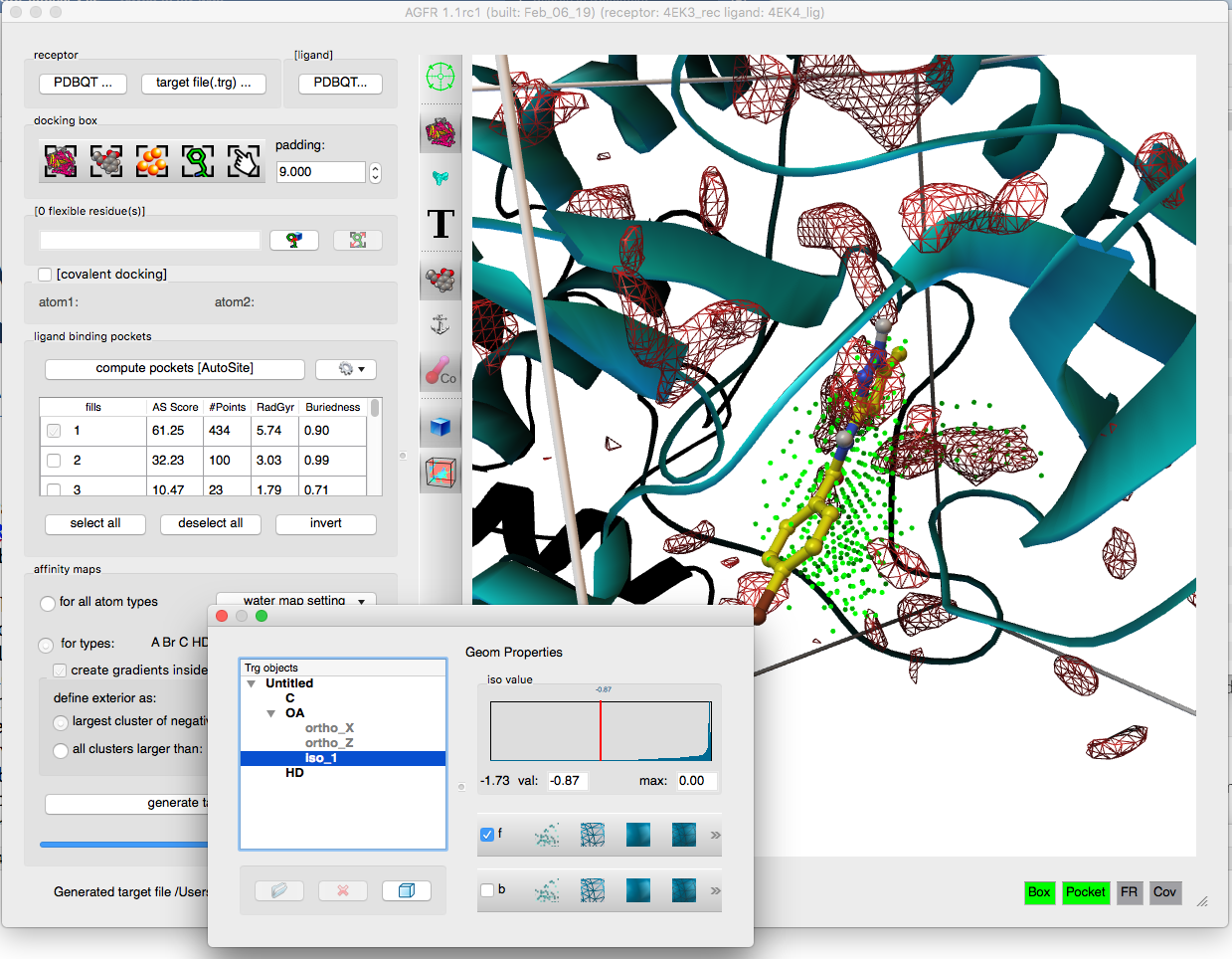
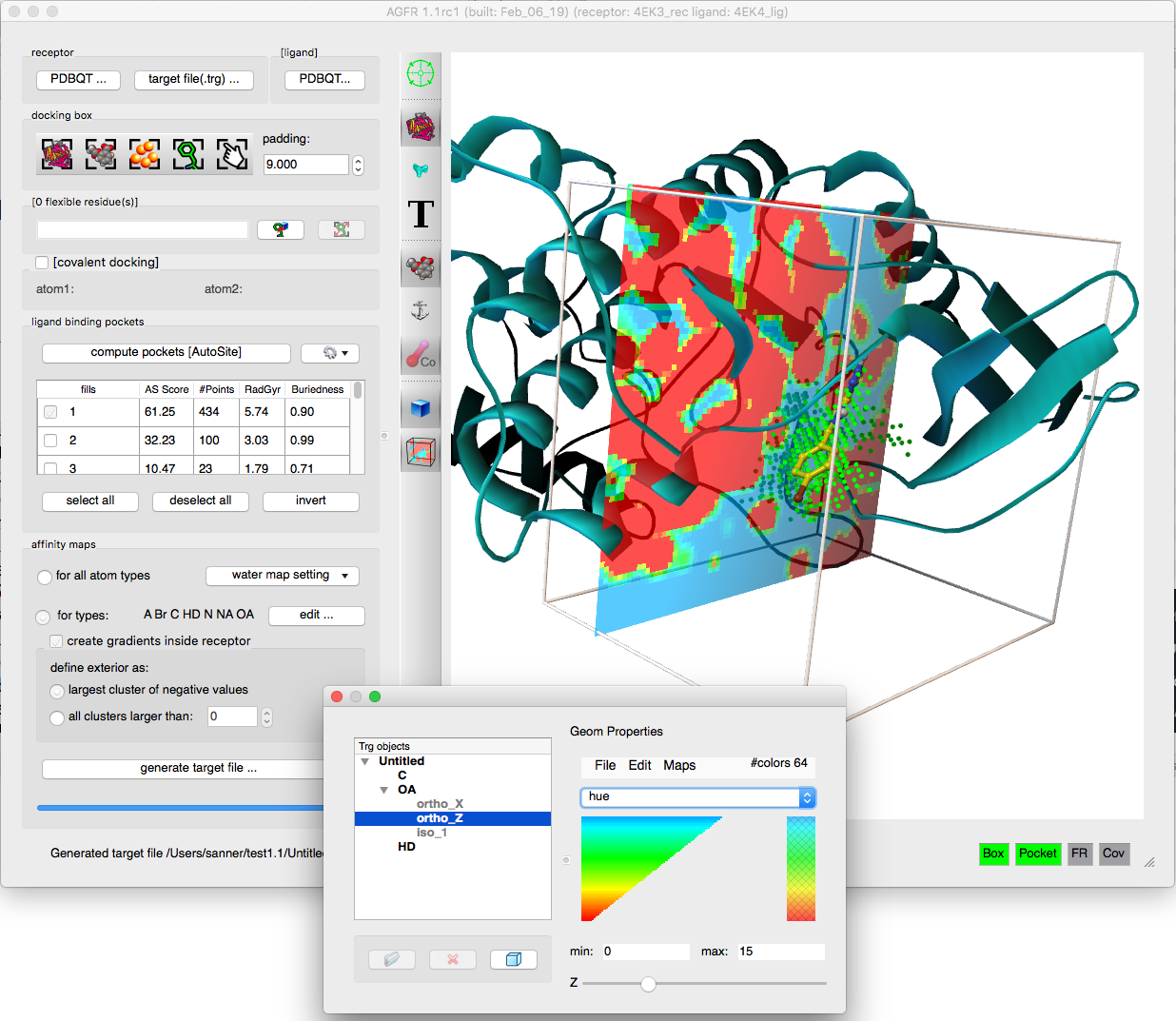
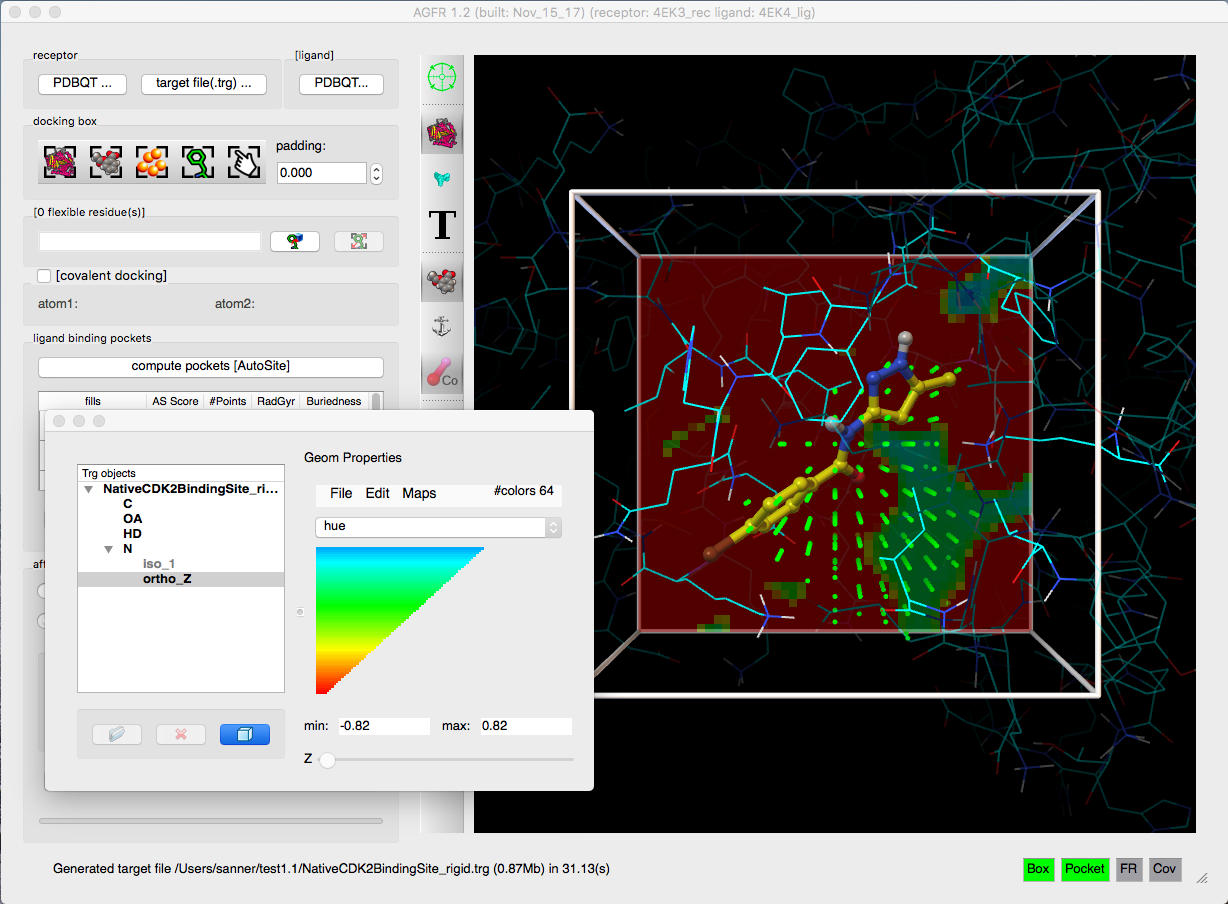
AGFR can be used to specify the docking box position and size in a variety of ways form the command line or using a graphical user interface. It uses AutoGrid4 to calculate the affinity maps, which are stored in “target files” (.trg extension) along with meta-data describing the maps and their calculation. The meta-data simplifies the management of maps and supports data provenance and reproducibility. Target files can be unzipped and the maps extracted for use with AutoDock4. We plan to add direct support for target files in the next release of AutoDock4.
Features:
- Calculation of affinity maps for:
- rigid receptors
- receptors with flexible side chains
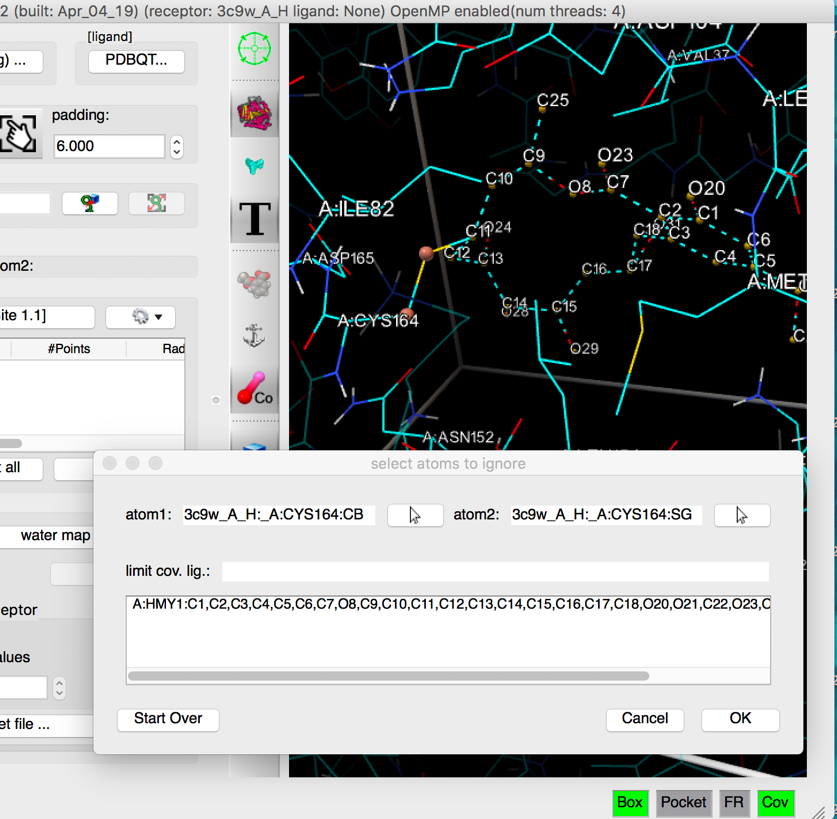
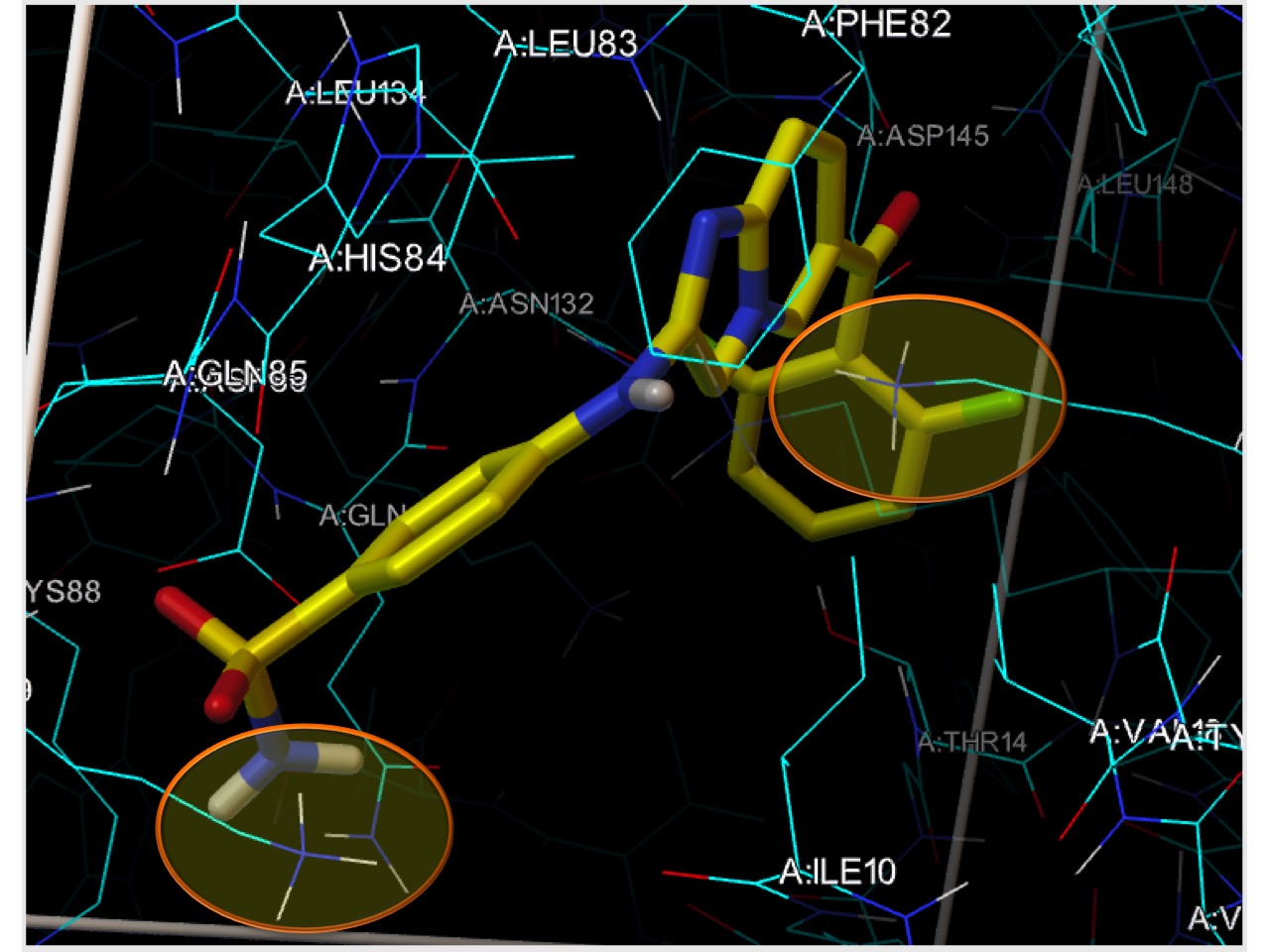
- covalent docking
- Generation of water maps for hydrated docking
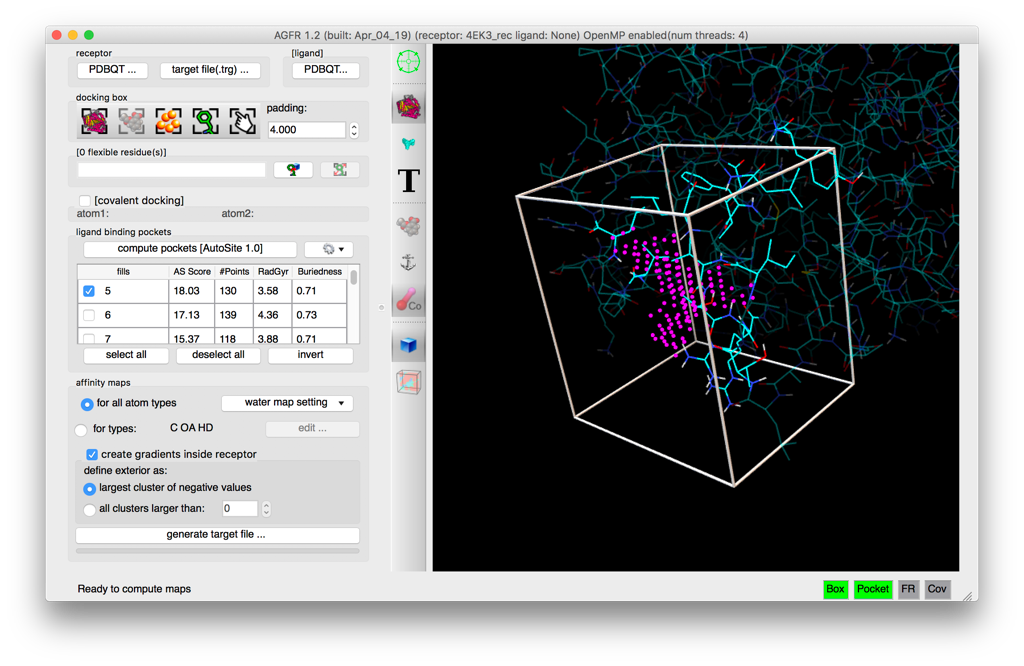
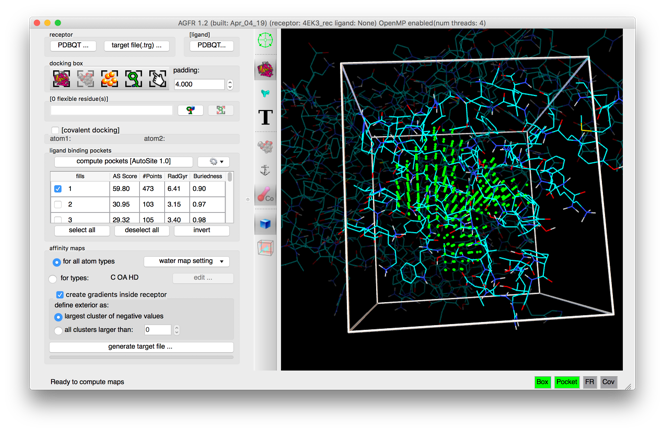
- Automatic binding pockets detection
- Addition of affinity gradients to the maps
- Modern graphical user interface
- Powerful command line
- Data provenance and reproducibility support