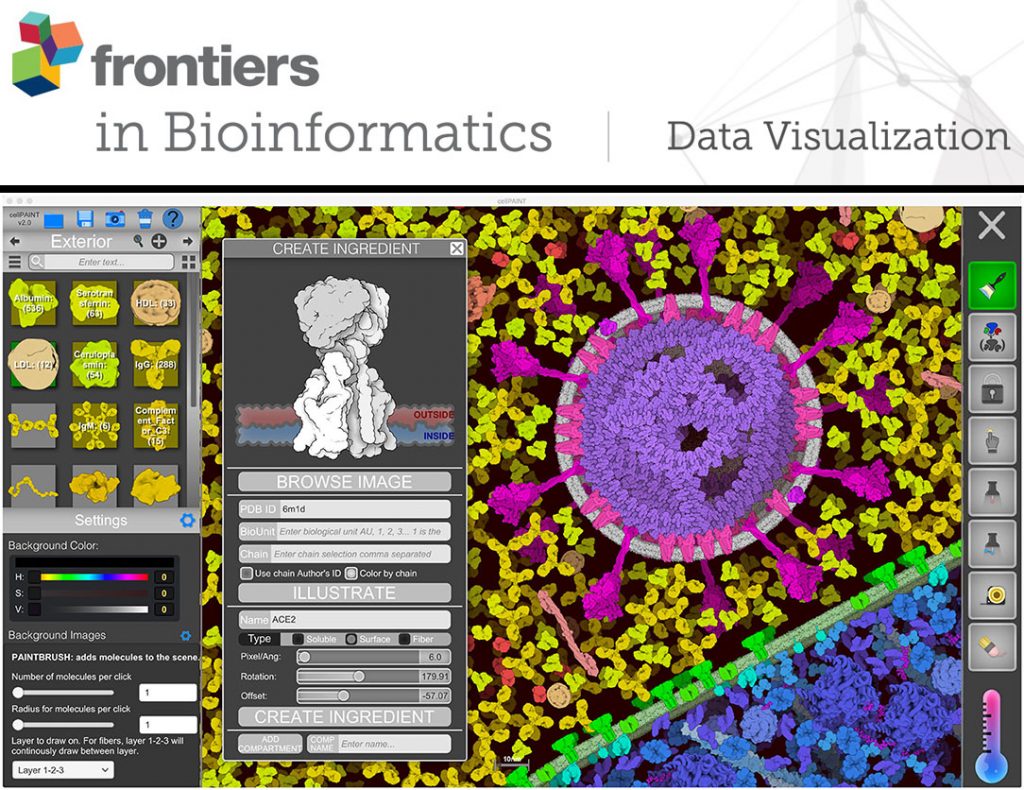
CellPAINT: Digital Illustration from Atoms to Cells
With CellPAINT, you can create dynamic illustrations of the inner molecular structure of viruses and living cells. CellPAINT pictures are based on experimental results from many different laboratories. The size and shape of each molecule is based on atomic structures determined by x-ray crystallography, NMR spectroscopy and electron microscopy, and key interactions, such as the embedding of proteins in membranes, are enforced as you create your pictures.
Our new release, CellPAINT-2.0, is now available. It allows you to import new molecules into the painting palette, and adds many options for refining your molecular paintings. In addition, we recently added some new molecules for creating illustrations of mRNA vaccines (CellPAINT-2.0.3). The web version is available below, and you can download a stand-alone version from SourceForge.
Please note: this is new software, so we are actively looking for people to test the program, share what they have created, and to give us feedback on usability and problems. There are a few Frequently-Asked Questions on the Documentation page. If you’d like to make a bug report, please use the “Tickets” at the SourceForge page.
Click here for a tutorial and documentation for CellPAINT-2.0.
Previous versions, such as CellPAINT-coronavirus, are available from the side menu.
Both the stand-alone and web versions of CellPAINT are free for use–please feel free to use the images that you create in any way that you like, including in commercial applications.
For users of the web version:
There was a small problem with saving and loading files, which is hopefully corrected now (the web version only supports zip files, and previously zip files were only written if you had created an ingredient). Also, the save/load buttons are not always responsive–sometimes you need to click again a second time anywhere in the program to get the dialog box. We’re trying to sort that out now.
The program is being developed by Adam Gardner, Ludovic Autin, Arthur Olson and David Goodsell, with support from the US National Institutes of Health. Thanks also to Martina Maritan for scientific support, and Melanie Stegman, Brett Barbaro and other members of the CCSB team for help with testing and usability analysis.
Our first paper describing CellPAINT is available on PubMedCentral PMC6456043.
Tell us what you think!
CellPAINT is currently in active development, and we are looking for testers. The software still has a few idiosyncracies, so please be prepared to restart the programs if you encounter problems. We are looking for feedback on several general questions:
Were you able to create the scenes that you wanted to make?
Will this type of program be useful in your own work?
Are there any features that you would like to see added?
Please send your impressions to Ludovic Autin at autin@scripps.edu.