This page was last updated on April 25th, 2019 at 11:40 pm
AutoGridFR overview
AutoGridFR (AGFR) is a software program for generating target files, i.e. a single file defining a binding site targeted for docking on a biological macro-molecule. Target files simplifies the management of maps and support data provenance and reproducibility. They can be unzipped and the maps extracted for use with AutoDock4. We plan to add direct support for target files in the next release of AutoDock4.
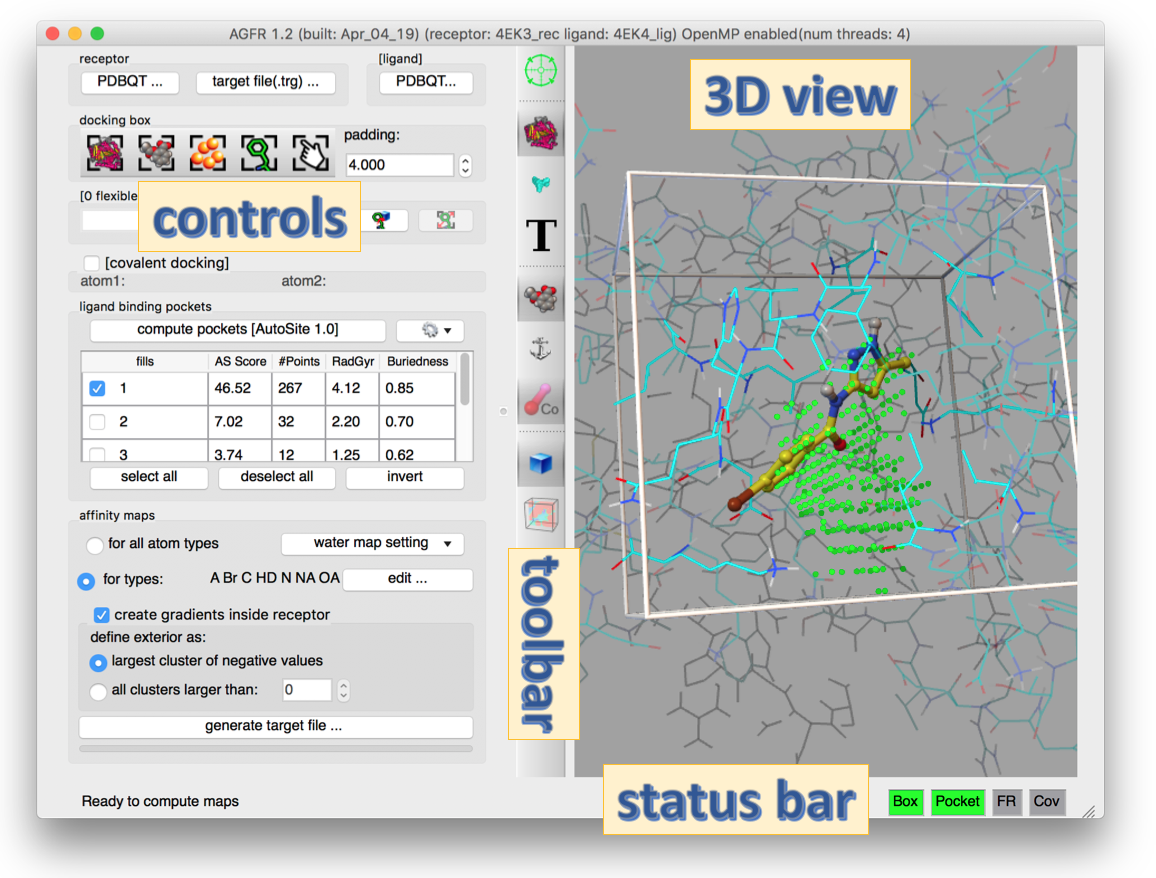
AGFR can be run form the command line of through a graphical user interface show in the figure below.
For the impatient you can skip the the use cases tutorials, or read on for a brief overview of the GUI.
The title of the GUI window displays the program version along with the names of the currently loaded receptor and ligand. In addition, if OpenMP is enabled, the number of hardware threads available will appear in the title.
The GUI has 4 main areas: 1) On the left, the “controls” panel provides vertically stacked boxed containing widgets for performing the tasks comprised in the workflow of computing target files for a given receptor; 2) On the right side a 3D view provides the user with real time visual feed back; 3) In the middle a toolbar presents buttons for altering the visual representation in the 3D view; and finally, 4) at the bottom, a status bar provides textual feedback about computations being performed or the next possible step in the workflow, as well as status lights (red or green) informing the user about that validity of the current docking box definition.
Below we give a quick overview of the function of the widgets in the control panel.
The input boxes entitled “receptor” and “ligand” are located at the top. They allows loading a receptor in the PDBQT file format, or open an existing target file which will configure all the widget according to the way the target file was generated. Optionally a crystallographic ligand can be loaded to help the user position the docking box and/or define the list of atom types for which to compute affinity maps.
Below, the widget “docking box” presents a group of buttons to set up the docking box (i.e. the rectilinear region of space for over which affinity maps will be computed). The choices are to size and position the box to encompass: the entire receptor, the ligand (enabled if one is loaded), a given pocket (enabled if pockets have been identified, see the “ligand binding pockets” section below), a user selected set of receptor amino acids, or by manually specify the position and size of the box, respectively. Any of these buttons (except the manual specification of the box) will calculate the bounding box corresponding to the pressed button and add the current padding values to each side of the bounding box. The box needs to overlap with the receptor for the BOX status light in the status bar to turn red.
Optionally, flexible receptor side chains can be specified using the third group of widgets “[0 flexible residue(s)]“. A selection string can be types or residues can be selected in a tree-widget. The syntax for a selection string is a chain ID followed by a colon an a comma separated list of residues, e.g. A:LYS33,LYS89;B:ARG12. The docking box is required to cover flexible residues or else the FR status light in the status bar will be red. A button is provide to adjust the box size to fulfill this requirement. More details are given in the tutorial “Affinity maps for docking with flexible receptor side-chains“.
The widgets in the “covalent Docking” section allow to optionally specify 2 atoms forming a covalent bond between the receptor and the ligand. these atoms can be picked in in the 3D view or specified by typing in their full names (e.g. A:CYS164:SG). The covalent ligand (if present in the loaded receptor) will be displayed with dashed lines and can be edited by adding or removing atoms. More details are available in the “Affinity maps for covalent docking” tutorial.
“ligand binding pockets“: The table widget in this section lists the various pockets identified by AutoSite on the current docking box. Fills can be selected to participate in the definition of the binding pocket. Both the original version 1.0 as well as version 1.1 of the AutoSite algorithm are available. Version 1.1 is slower but provides better pocket descriptors, in particular for larger ligands such as peptides.
The final group of widgets (“affinity maps“) is used to specify the list of atom types for which affinity maps are computed. Maps can be computed for all known AutoDock atom types which is the preferred option as it creates a target file usable for any ligand. AGFR also automatically computes the water maps for hydrated docking. The parameters associated with the calculation of water maps can be set here. Finally, the user can specify whether gradients are added to the maps, and specify options for their calculation.
Most buttons in the interface are only enabled once prerequisites for performing the associated task are met. For instance, the “generate target files …” button will only be enabled after a receptor is loaded, a box that overlaps with the receptor is defined, at least one fill defining a pocket is selected and the selected fill overlaps with the box. The 4 buttons at the bottom right, will be red initially, and turn green gradually as conditions become satisfied, thus guiding the user through the process.
The actual calculation of the maps is performed by an optimized, and OpenMP enable version of AutoGrid4, bundled with the application.
