Modeling and Visualizating the Cellular Mesoscale
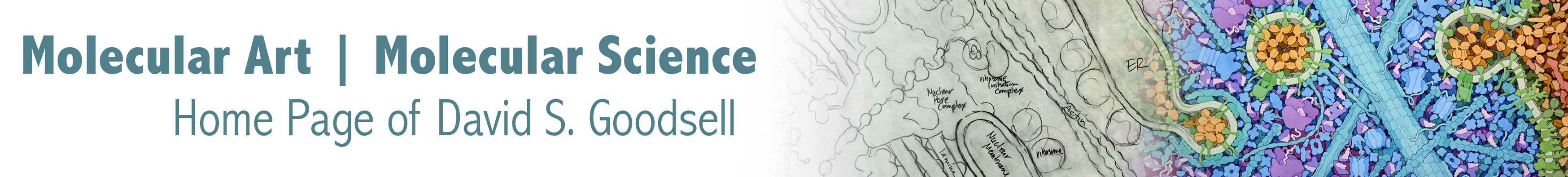
CellPACK combines information from cellular ultrastructure, genomics and proteomics, and atomic structure to build three-dimensional models of subcellular environments at molecular detail. In our current work, we are bringing cellPACK technology and the associated visualization tool cellVIEW to a new level with an ambitious goal: the modeling and visualization of an entire bacterial cell. The methods are being applied in two settings: as a tool for research in the study of bacterial structure and function, and in educational settings as a way to promote understanding of structure and function of living cells.
This work is currently supported by an R01 grant from the National Institutes of Health. To learn more about this and associated projects looking at the cellular mesoscale, take a look at the Mesoscale page at the CCSB site.
Automated Docking for Drug Discovery and Design
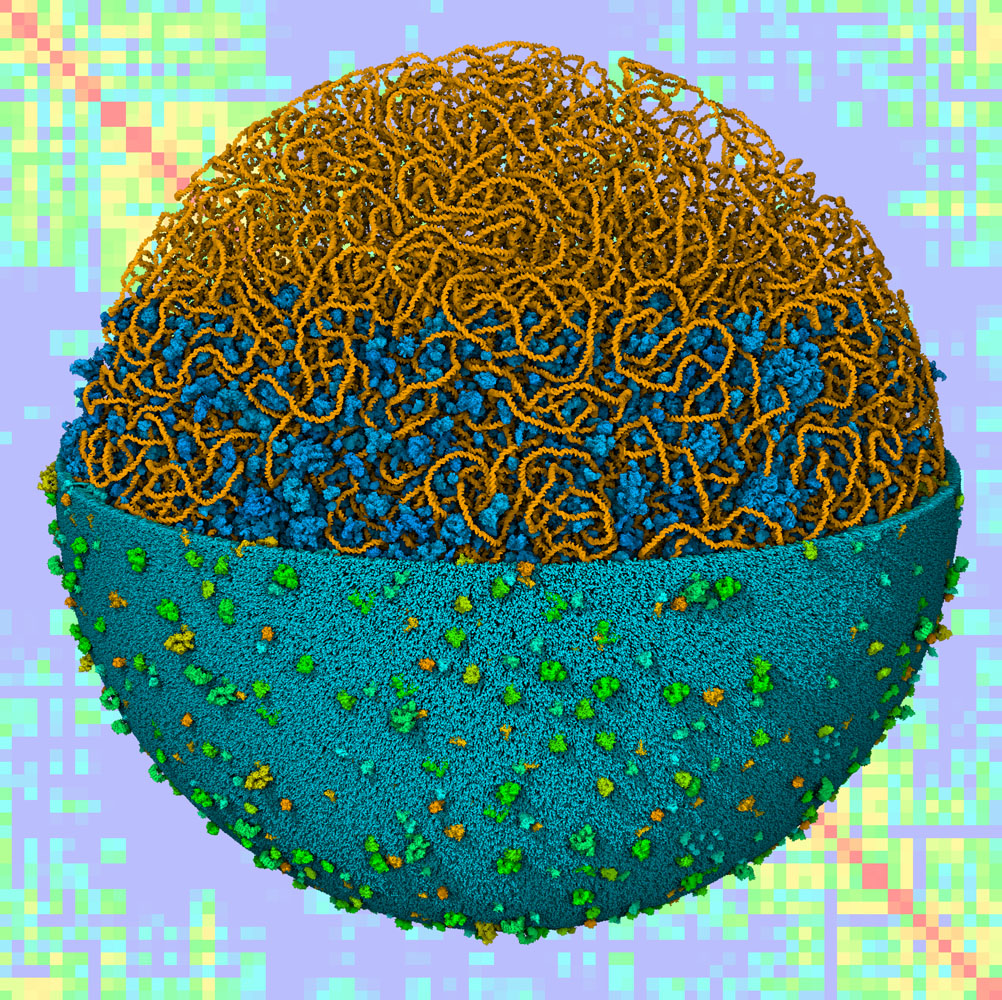
Computational docking and virtual screening are currently a central resource in the discovery and development of new drugs to fight the most challenging threats to our health. There are numerous examples of success, targeting receptors, enzymes, channels, nucleic acids, and many other targets. In 1990, AutoDock was the first method for computational docking that allowed flexibility in the ligand and used a detailed physics-based force field. Our continuing development to address the needs of the community, free accessibility of source code, documentation, and tutorials have placed AutoDock as the most cited docking method currently available.
This work is supported by an R01 grant from the National Institutes of Health (Stefano Forli, PI) for the continued development and maintenance of the AutoDock suite. For more information, visit the AutoDock page at the CCSB site.
HIV Biology
Members of the HIVE Center are characterizing at the atomic level the structural and dynamic relationships between interacting macromolecules in the HIV life cycle. We focus on interactions of the major HIV enzymes with their partners and effectors since they encompass key processes in the viral life cycle and as existing drug targets provide a rich base of structural, biological and evolutionary data that inform our goals. We explore resistance evolution in HIV as an opportune platform upon which to characterize the dynamic relationships between interacting macromolecular structures at the atomic level. Our approach is significant due to the promise of new structural insights into the interdependence of viral mechanisms and the direct potential for new drug design methodologies and therapeutic strategies.
Within the HIVE Center, I have focused on modeling the mesoscale properties of HIV at various stages of the viral lifecycle, and how inhibitors can block key viral processes. I have also created educational and outreach materials presenting the work of HIVE Center members. For more information, visit the HIVE Center site.